🌲 Introduction
Forest plots are essential visualization tools in biomedical research, epidemiology, and statistical analysis. They excel at presenting:
- Regression analysis results (Logistic, Cox proportional hazards)
- Meta-analysis (combining multiple studies)
- Subgroup comparisons (stratified analyses)
A well-designed forest plot not only conveys statistical results clearly but also meets publication standards for journals and conferences.
Why evanverse?
While R has several packages for creating forest plots (e.g.,
forestplot, forestploter, meta),
evanverse provides:
✅ Publication-ready defaults - Beautiful themes out of the box ✅ Comprehensive customization - 40+ parameters for fine control ✅ Multi-model comparison - Compare multiple regression models side-by-side ✅ Intelligent automation - Auto-formatting, alignment, and significance highlighting ✅ Batch processing - Efficient workflows for large-scale analyses
This vignette demonstrates how to create professional forest plots from basic examples to advanced customizations.
📊 Understanding Forest Plots
What is a Forest Plot?
A forest plot displays effect estimates and their confidence intervals for multiple variables or studies. Each row represents:
- Variable name - The predictor or study identifier
- Point estimate - Effect size (OR, HR, RR, MD, etc.)
- Confidence interval - Uncertainty range (typically 95% CI)
- P-value - Statistical significance indicator
- Reference line - No-effect threshold (OR/HR = 1, MD = 0)
Key Components
┌─────────────┬────────────┬───────────┬─────────┐
│ Variable │ CI Plot │ OR (CI) │ P-value │
├─────────────┼────────────┼───────────┼─────────┤
│ Age ≥65 │ ──■── │ 1.45 (...)│ 0.001 │
│ Male │ ──■── │ 0.88 (...)│ 0.189 │
│ Smoking │ ──■── │ 1.67 (...)│ <0.001 │
└─────────────┴────────────┴───────────┴─────────┘
↑ ↑
Reference Estimate
line & CI box🚀 Quick Start
Installation
# Install from CRAN (when available)
install.packages("evanverse")
# Or install development version from GitHub
# install.packages("devtools")
devtools::install_github("evanbio/evanverse")Minimal Example (5 minutes)
Let’s create a basic forest plot using the built-in
forest_data dataset.
Step 1: Load and Inspect Data
# Load built-in example data
data("forest_data")
# Inspect structure
head(forest_data, 10)
#> variable est lower upper pval est_2 lower_2 upper_2 pval_2 est_3
#> 1 Age 1.28 1.15 1.43 0.001 NA NA NA NA NA
#> 2 BMI 1.05 1.02 1.09 0.003 NA NA NA NA NA
#> 3 Sex NA NA NA NA NA NA NA NA NA
#> 4 Male 1.42 1.10 1.83 0.007 NA NA NA NA NA
#> 5 Female 0.88 0.65 1.18 0.380 NA NA NA NA NA
#> 6 BMI category NA NA NA NA NA NA NA NA NA
#> 7 <25 1.00 0.75 1.33 1.000 NA NA NA NA NA
#> 8 25-30 1.35 0.95 1.92 0.095 NA NA NA NA NA
#> 9 ≥30 1.78 1.25 2.53 0.002 NA NA NA NA NA
#> 10 Smoking NA NA NA NA NA NA NA NA NA
#> lower_3 upper_3 pval_3 n_total n_event event_pct color_id note
#> 1 NA NA NA 850 210 24.7 cont <NA>
#> 2 NA NA NA 850 210 24.7 cont <NA>
#> 3 NA NA NA NA NA NA <NA> <NA>
#> 4 NA NA NA 425 120 28.2 sex <NA>
#> 5 NA NA NA 425 90 21.2 sex <NA>
#> 6 NA NA NA NA NA NA <NA> <NA>
#> 7 NA NA NA 320 55 17.2 bmi <NA>
#> 8 NA NA NA 310 72 23.2 bmi <NA>
#> 9 NA NA NA 220 83 37.7 bmi <NA>
#> 10 NA NA NA NA NA NA <NA> <NA>Step 2: Prepare Display Data
The key to plot_forest() is preparing a display
data frame with all text columns you want to show.
# Filter single-model data
df_single <- forest_data %>%
filter(is.na(est_2)) %>% # Single model (no est_2)
filter(!is.na(est)) %>% # Remove header rows
head(10) # First 10 rows for demo
# Create display table
plot_data <- df_single %>%
mutate(
` ` = strrep(" ", 20), # Blank column for CI graphic
`OR (95% CI)` = sprintf("%.2f (%.2f-%.2f)", est, lower, upper),
`P` = ifelse(pval < 0.001, "<0.001", sprintf("%.3f", pval)),
`N` = n_total
) %>%
select(Variable = variable, ` `, `OR (95% CI)`, `P`, `N`)
print(plot_data)
#> Variable OR (95% CI) P N
#> 1 Age 1.28 (1.15-1.43) 0.001 850
#> 2 BMI 1.05 (1.02-1.09) 0.003 850
#> 3 Male 1.42 (1.10-1.83) 0.007 425
#> 4 Female 0.88 (0.65-1.18) 0.380 425
#> 5 <25 1.00 (0.75-1.33) 1.000 320
#> 6 25-30 1.35 (0.95-1.92) 0.095 310
#> 7 ≥30 1.78 (1.25-2.53) 0.002 220
#> 8 Current/Former 1.52 (1.18-1.96) 0.001 380
#> 9 Control 1.00 (0.80-1.25) 1.000 280
#> 10 Treatment A 0.68 (0.48-0.96) 0.028 285Step 3: Create Forest Plot
# Create forest plot
p1 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2, # Column for CI graphic (blank column)
ref_line = 1, # OR = 1 reference
xlim = c(0.5, 2.5),
arrow_lab = c("Lower Risk", "Higher Risk")
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 40, 35, 40, 20, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 6.1, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p1)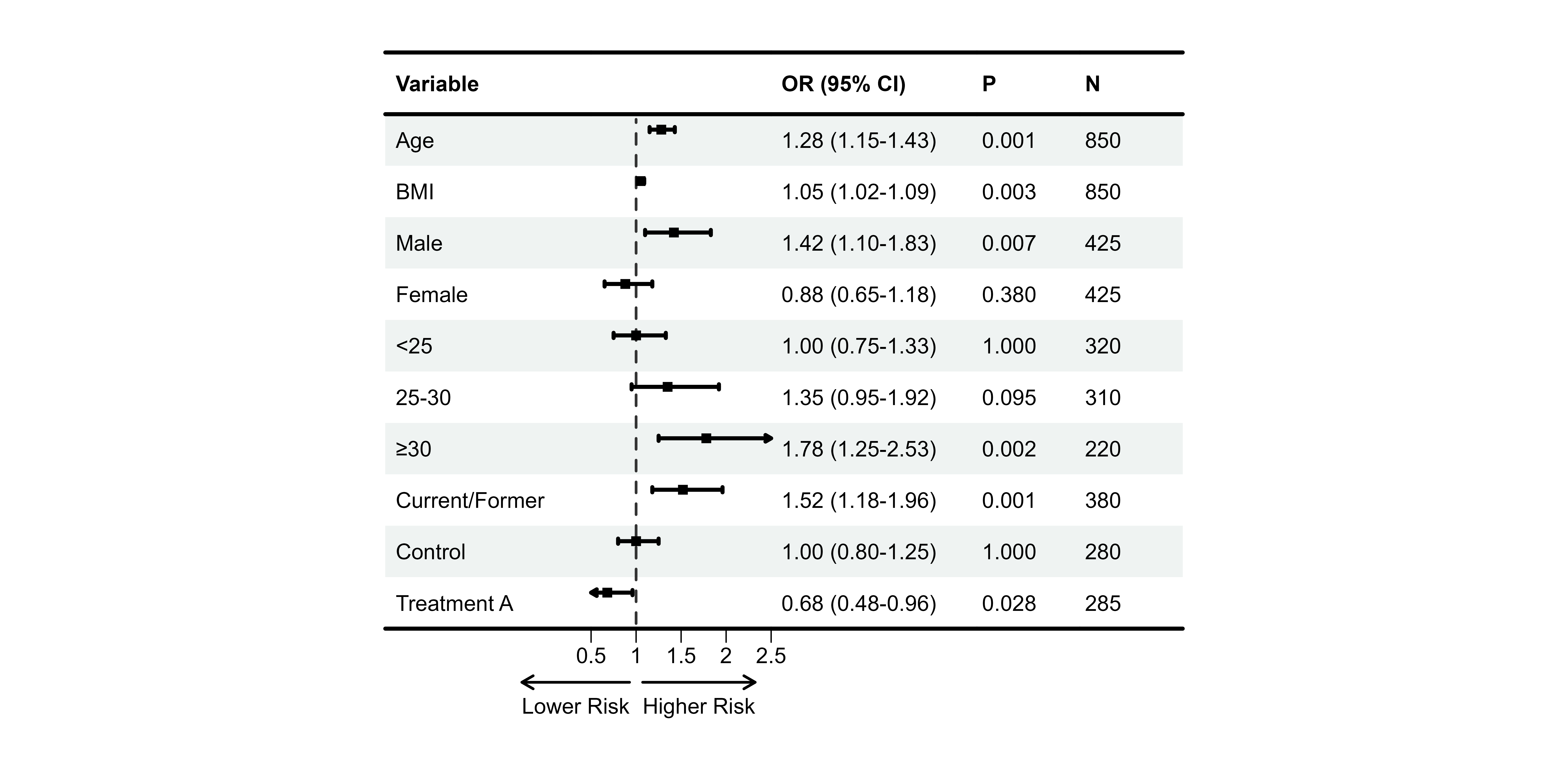
🎉 Congratulations! You just created your first publication-ready forest plot.
🎨 Single-Model Forest Plots
Now let’s explore how to customize single-model forest plots for different scenarios.
Data Preparation Deep Dive
Understanding the data structure is crucial for
plot_forest():
# YOUR data frame should have:
# 1. Display columns (text, formatted strings)
# 2. Numeric vectors for est, lower, upper (NOT in data frame)
# 3. A blank column (" ") where CI graphics will be drawn
plot_data <- data.frame(
Variable = c("Age", "Sex", "BMI"), # Display
` ` = rep(strrep(" ", 20), 3), # Blank for CI
`OR (95% CI)` = c("1.45 (...)", ...), # Display
`P` = c("0.001", "0.189", "0.045") # Display
)
# Numeric vectors (not in data frame)
est_values <- c(1.45, 0.88, 1.35)
lower_values <- c(1.10, 0.65, 1.05)
upper_values <- c(1.83, 1.18, 1.71)Key Points: - Display data and numeric data are
separate - Use sprintf() to format OR/HR
strings - Create blank column with strrep(" ", width)
Theme Customization
Using Preset Themes
# Default theme (built-in)
p2 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
theme_preset = "default"
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 40, 35, 40, 20, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p2)
Custom Theme Parameters
# Override specific theme parameters
p3 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
theme_custom = list(
base_size = 14, # Larger font
ci_pch = 18, # Diamond shape
ci_lwd = 2, # Thicker lines
ci_fill = "#4DBBD5", # Custom color
ci_Theight = 0.15 # Box height
)
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 37.8, 31.4, 39.1, 16.4, 12.2, 5
#> Adjusted: 10, 45, 40, 45, 25, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7.5, 6.5, 6.5, 6.5, 6.5, 6.5, 6.5, 6.5, 6.5, 6.5, 6.5, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p3)
Text Alignment
Professional tables require proper alignment:
p4 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
align_left = 1, # Variable names left
align_center = c(2, 3), # CI column and OR center
align_right = c(4, 5) # P-value and N right
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 40, 35, 40, 20, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p4)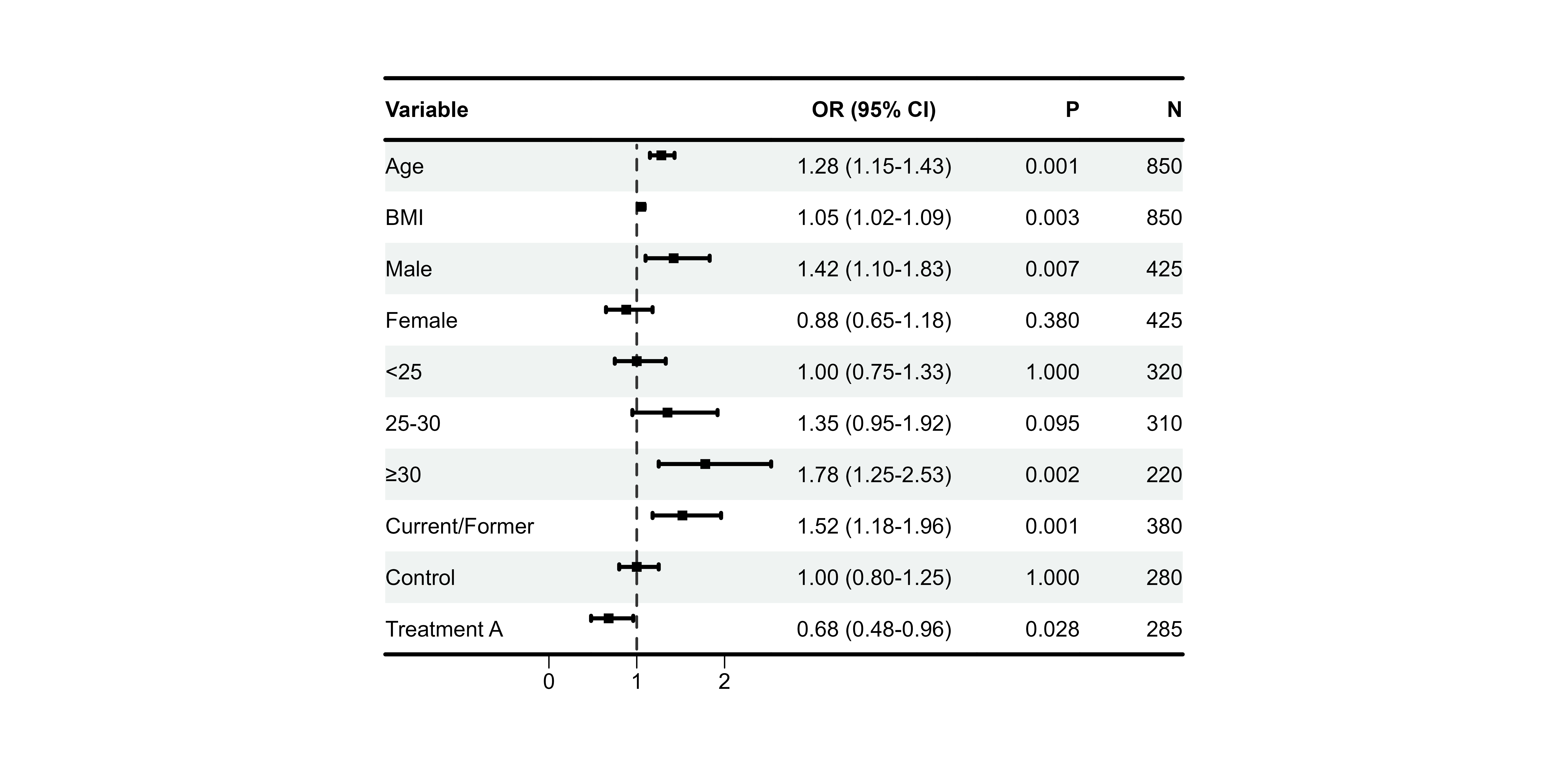
Bold Formatting
Highlighting Group Headers
# Assuming "Sex" and "BMI category" are group headers
p5 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
bold_group = c("Sex", "BMI category"),
bold_group_col = 1
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 40, 35, 40, 20, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p5)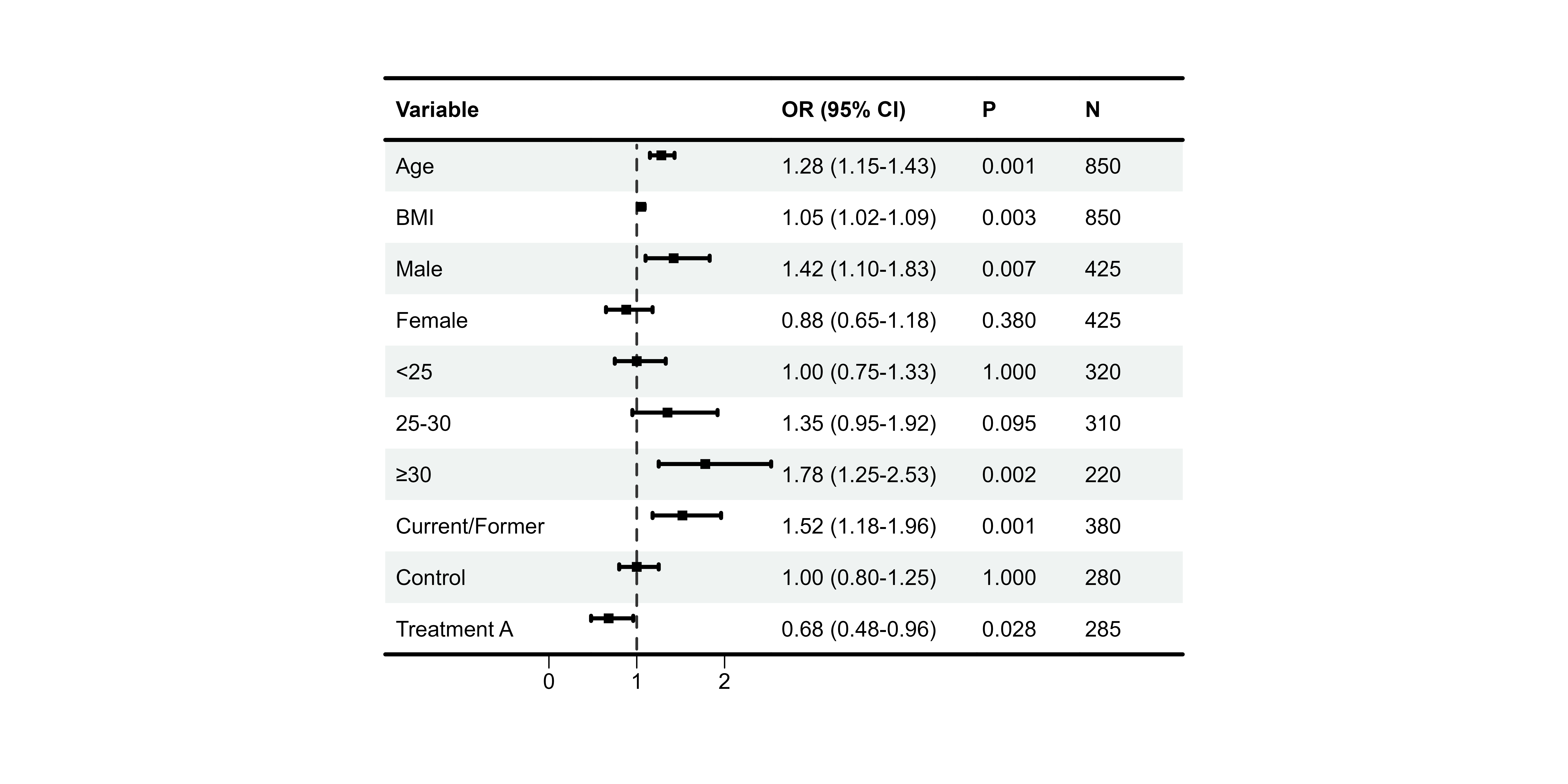
Auto-Bold Significant P-values
p6 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
bold_pvalue_cols = 4, # P-value column
p_threshold = 0.05 # Significance level
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 40, 35, 40, 20, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p6)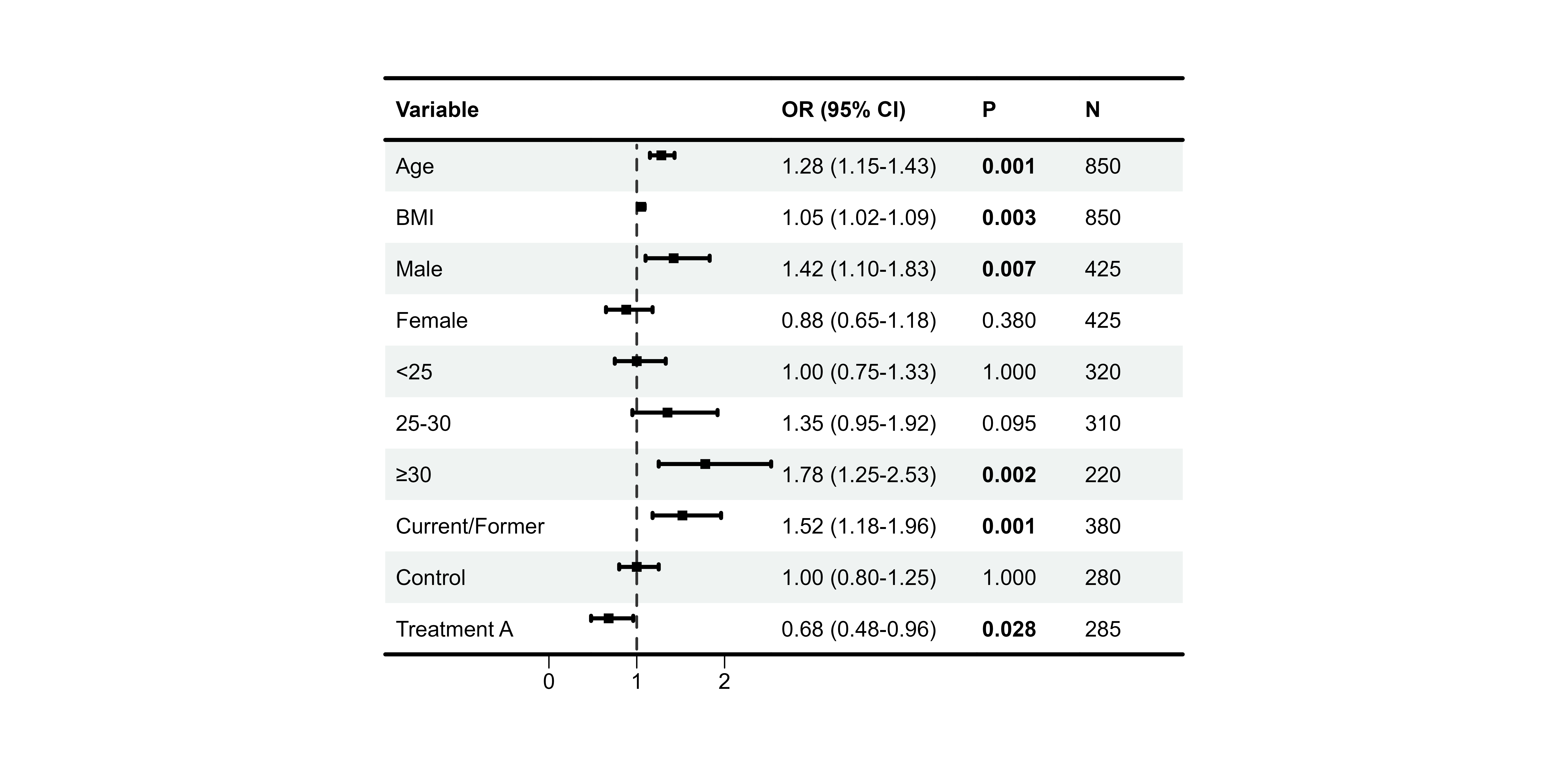
Background Styles
Zebra Stripes
p7 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
background_style = "zebra",
background_colors = list(
primary = "#F0F0F0",
secondary = "white"
)
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 40, 35, 40, 20, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p7)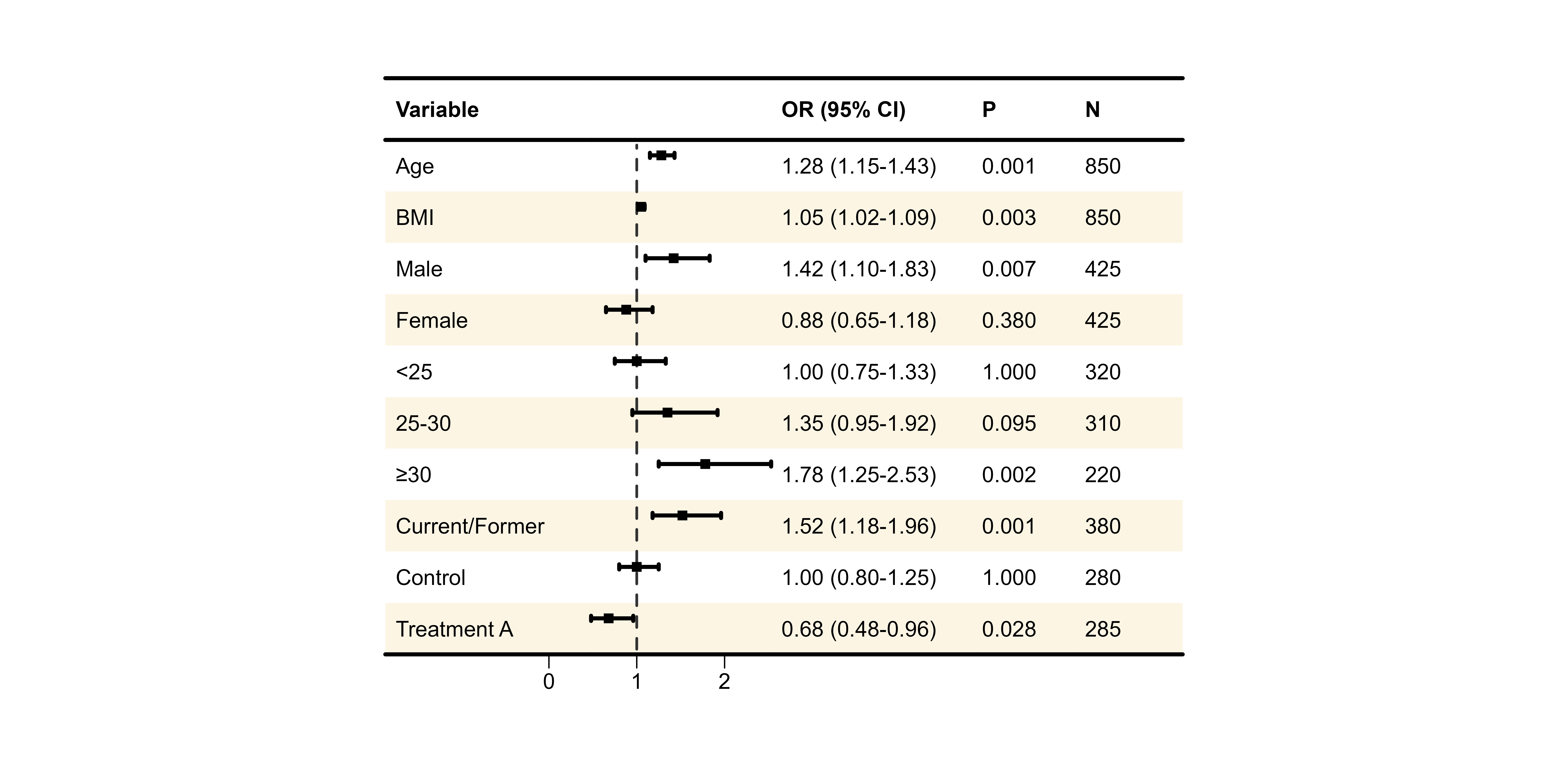
Group-based Coloring
# Identify rows that are group headers (NA in est)
group_rows <- which(is.na(df_single$est))
p8 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
background_style = "group",
background_group_rows = group_rows,
background_colors = list(
primary = "#E3F2FD", # Group headers
secondary = "white" # Data rows
)
)
#> Warning: Style 'group' requires `group_rows`. No background applied.
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 40, 35, 40, 20, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p8)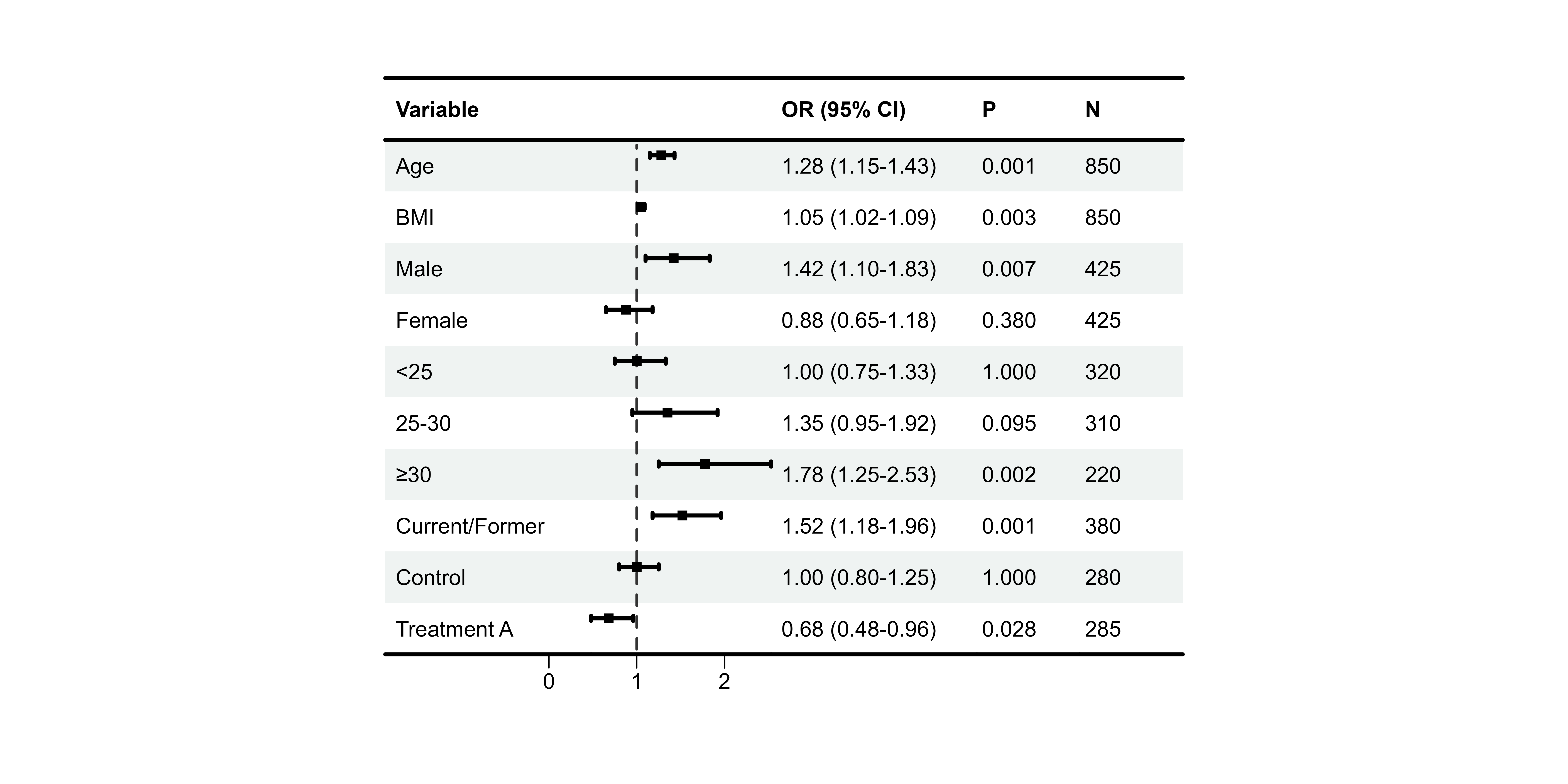
CI Colors
Single Color
p9 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
ci_colors = "#E64B35" # All boxes same color
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 40, 35, 40, 20, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p9)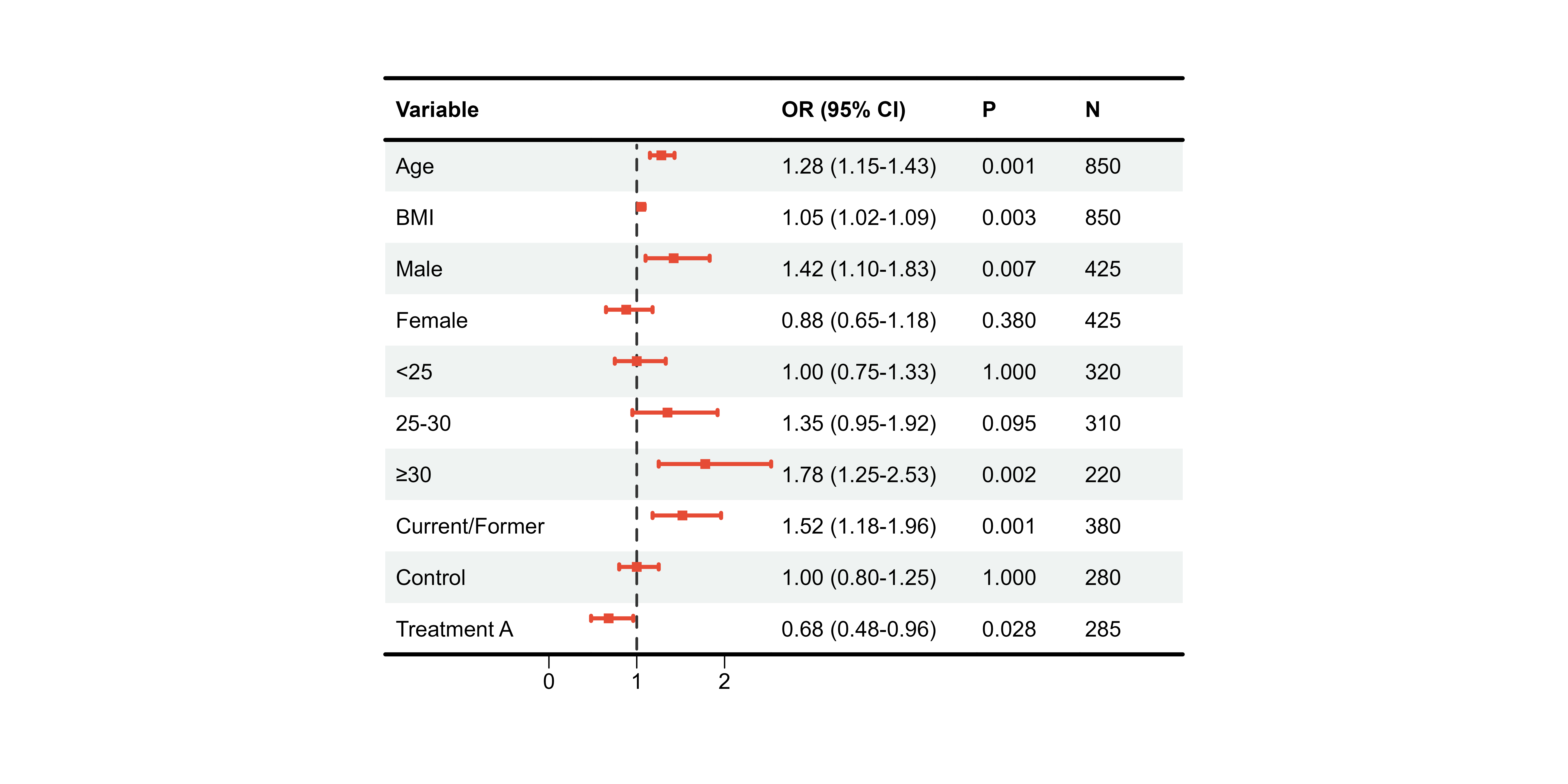
Color by Significance
# Color based on p-value
ci_cols <- ifelse(df_single$pval < 0.05, "#E64B35", "#CCCCCC")
p10 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
ci_colors = ci_cols # Vector matching rows
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 40, 35, 40, 20, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p10)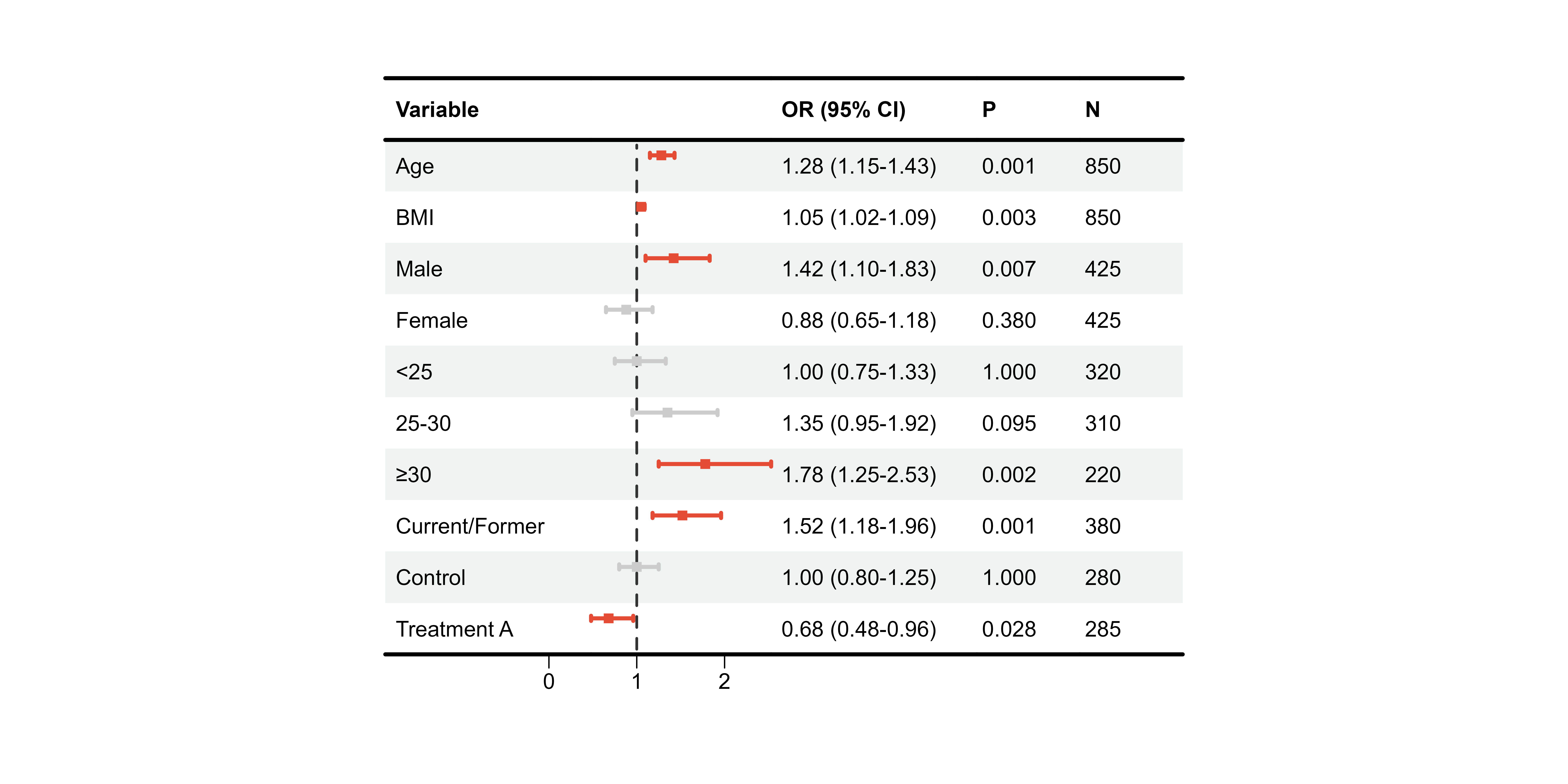
Borders
p11 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
add_borders = TRUE,
border_width = 3
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 40, 35, 40, 20, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p11)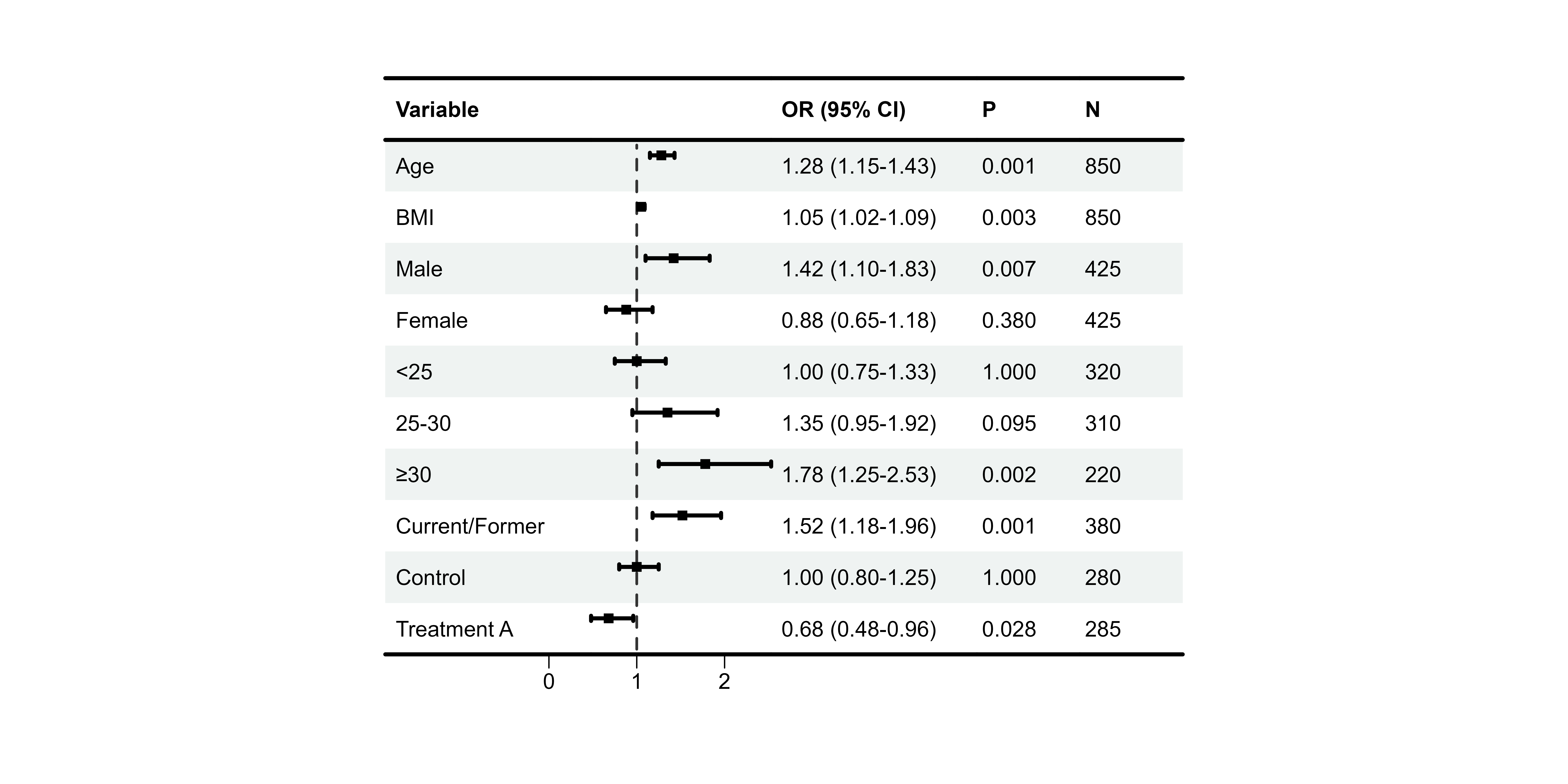
Complete Customization Example
# All customizations combined
p12 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
xlim = c(0.5, 2.5),
arrow_lab = c("Protective", "Risk"),
# Alignment
align_left = 1,
align_center = c(2, 3),
align_right = c(4, 5),
# Bold
bold_pvalue_cols = 4,
p_threshold = 0.05,
# Background
background_style = "zebra",
# CI colors by significance
ci_colors = ifelse(df_single$pval < 0.05, "#E64B35", "#4DBBD5"),
# Borders
add_borders = TRUE,
# Layout
height_main = 10,
height_bottom = 8,
layout_verbose = FALSE
)
print(p12)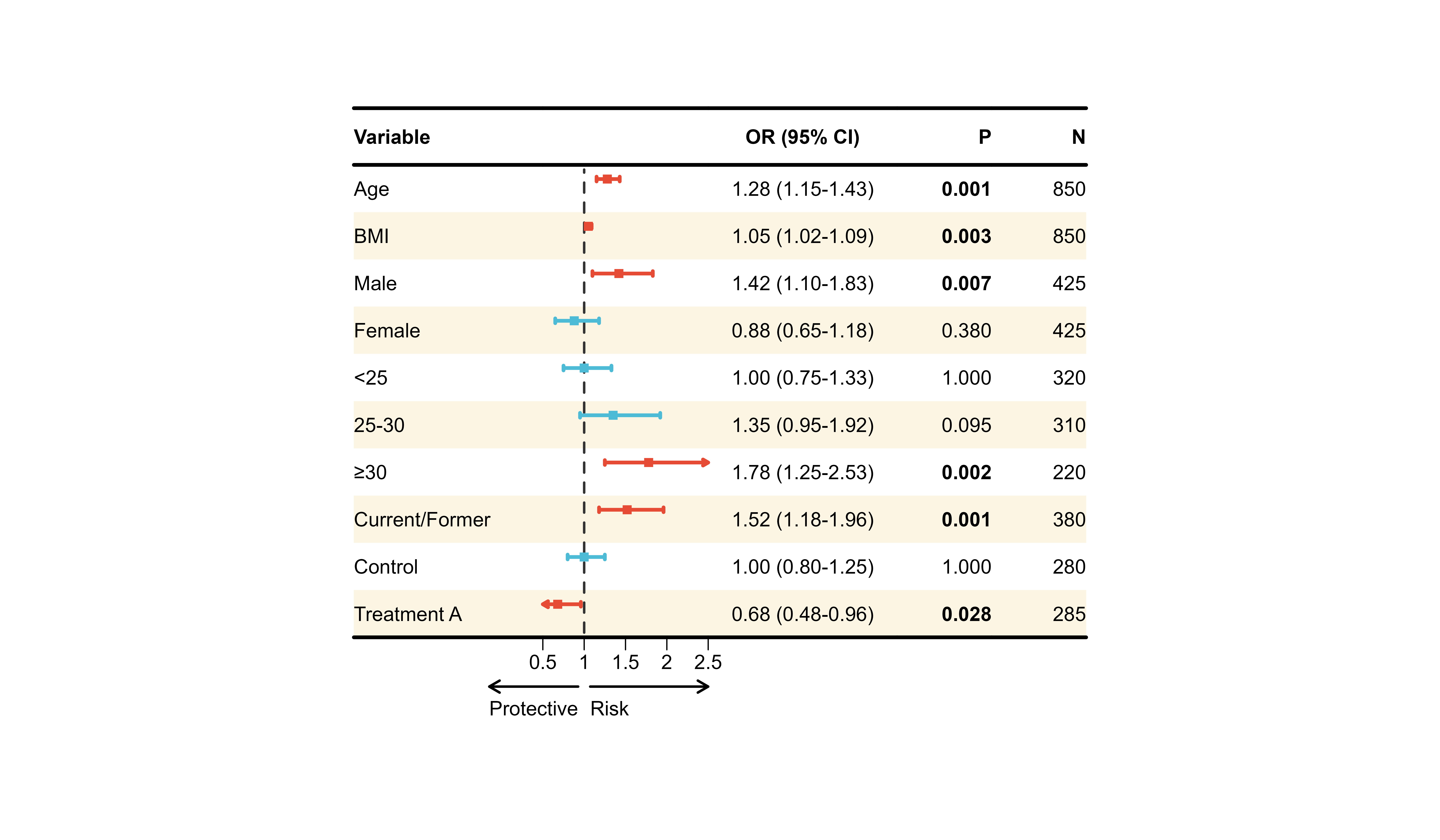
🔄 Multi-Model Comparison
One powerful feature of plot_forest() is comparing
multiple regression models side-by-side.
Preparing Multi-Model Data
# Filter multi-model data
df_multi <- forest_data %>%
filter(!is.na(est_2)) # Has multiple models
# Create display table with multiple model columns
plot_data_multi <- df_multi %>%
mutate(
` ` = strrep(" ", 15),
`Model 1` = sprintf("%.2f (%.2f-%.2f)", est, lower, upper),
`Model 2` = sprintf("%.2f (%.2f-%.2f)", est_2, lower_2, upper_2),
`Model 3` = sprintf("%.2f (%.2f-%.2f)", est_3, lower_3, upper_3)
) %>%
select(Variable = variable, ` `, `Model 1`, `Model 2`, `Model 3`)
print(plot_data_multi)
#> Variable Model 1 Model 2
#> 1 Hypertension 1.85 (1.42-2.41) 1.72 (1.32-2.24)
#> 2 Diabetes 2.12 (1.58-2.84) 1.95 (1.45-2.62)
#> 3 Physical inactivity 1.42 (1.08-1.87) 1.35 (1.03-1.77)
#> Model 3
#> 1 1.58 (1.20-2.08)
#> 2 1.78 (1.31-2.42)
#> 3 1.28 (0.97-1.69)Basic Multi-Model Plot
p13 <- plot_forest(
data = plot_data_multi,
est = list(df_multi$est, df_multi$est_2, df_multi$est_3),
lower = list(df_multi$lower, df_multi$lower_2, df_multi$lower_3),
upper = list(df_multi$upper, df_multi$upper_2, df_multi$upper_3),
ci_column = 2,
ref_line = 1,
xlim = c(0.5, 3)
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 37.2, 21.6, 34.1, 34.1, 34.1, 22.3, 5
#> Adjusted: 10, 45, 30, 40, 40, 40, 30, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p13)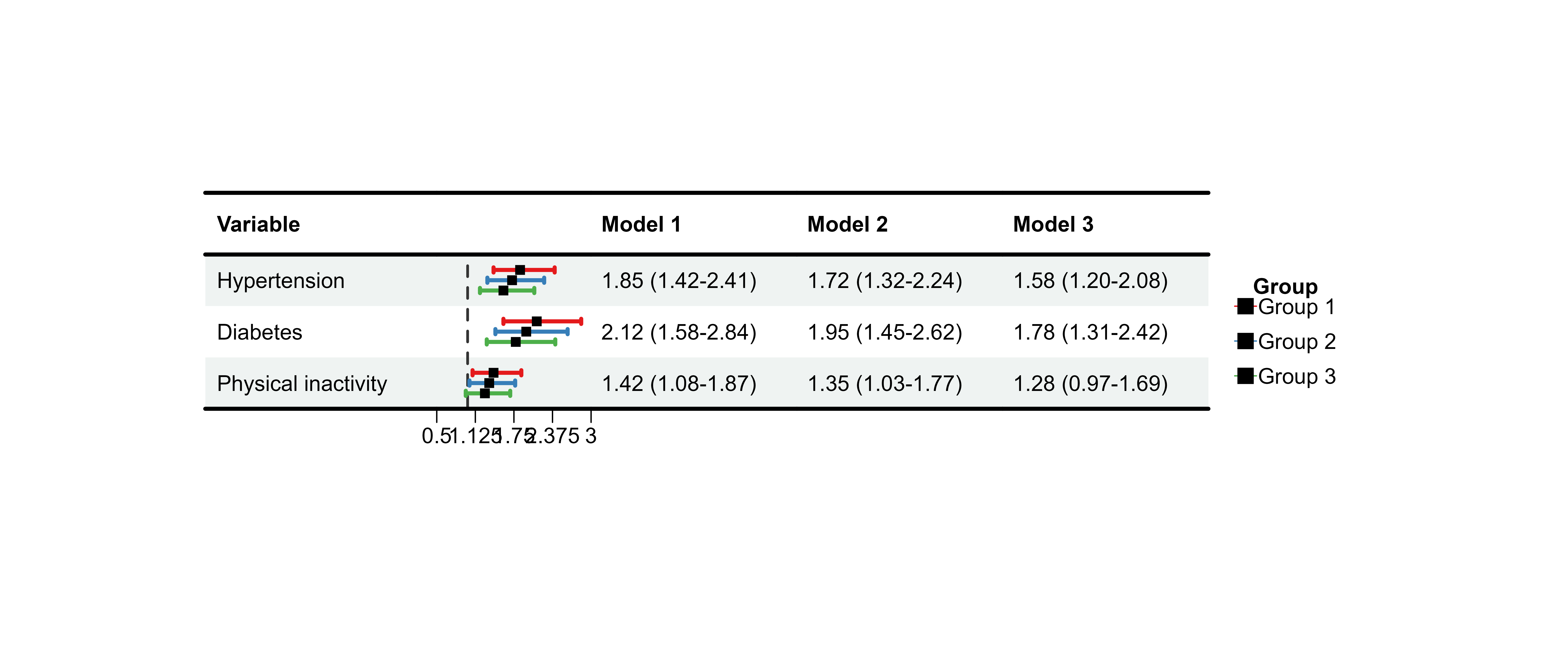
Adjusting Multi-Model Spacing
Use nudge_y to control vertical spacing between
models:
p14 <- plot_forest(
data = plot_data_multi,
est = list(df_multi$est, df_multi$est_2, df_multi$est_3),
lower = list(df_multi$lower, df_multi$lower_2, df_multi$lower_3),
upper = list(df_multi$upper, df_multi$upper_2, df_multi$upper_3),
ci_column = 2,
ref_line = 1,
xlim = c(0.5, 3),
nudge_y = 0.3 # Increase spacing
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 37.2, 21.6, 34.1, 34.1, 34.1, 22.3, 5
#> Adjusted: 10, 45, 30, 40, 40, 40, 30, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p14)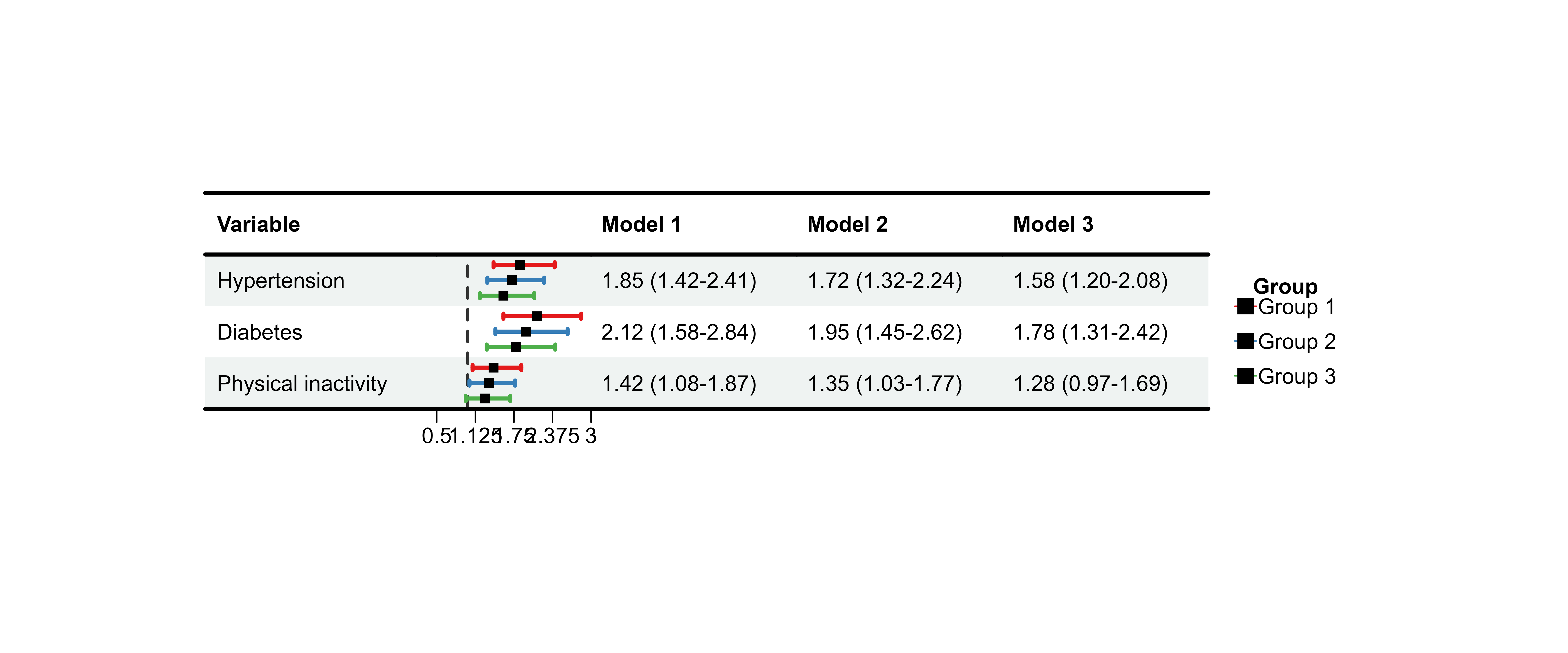
Customizing Model Appearance
Different Sizes per Model
# IMPORTANT: sizes must match number of ROWS, not models!
# For 3 rows, repeat the pattern
sizes_vec <- rep(0.6, nrow(plot_data_multi))
p15 <- plot_forest(
data = plot_data_multi,
est = list(df_multi$est, df_multi$est_2, df_multi$est_3),
lower = list(df_multi$lower, df_multi$lower_2, df_multi$lower_3),
upper = list(df_multi$upper, df_multi$upper_2, df_multi$upper_3),
ci_column = 2,
ref_line = 1,
xlim = c(0.5, 3),
sizes = sizes_vec # Must match row count!
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 37.2, 21.6, 34.1, 34.1, 34.1, 22.3, 5
#> Adjusted: 10, 45, 30, 40, 40, 40, 30, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p15)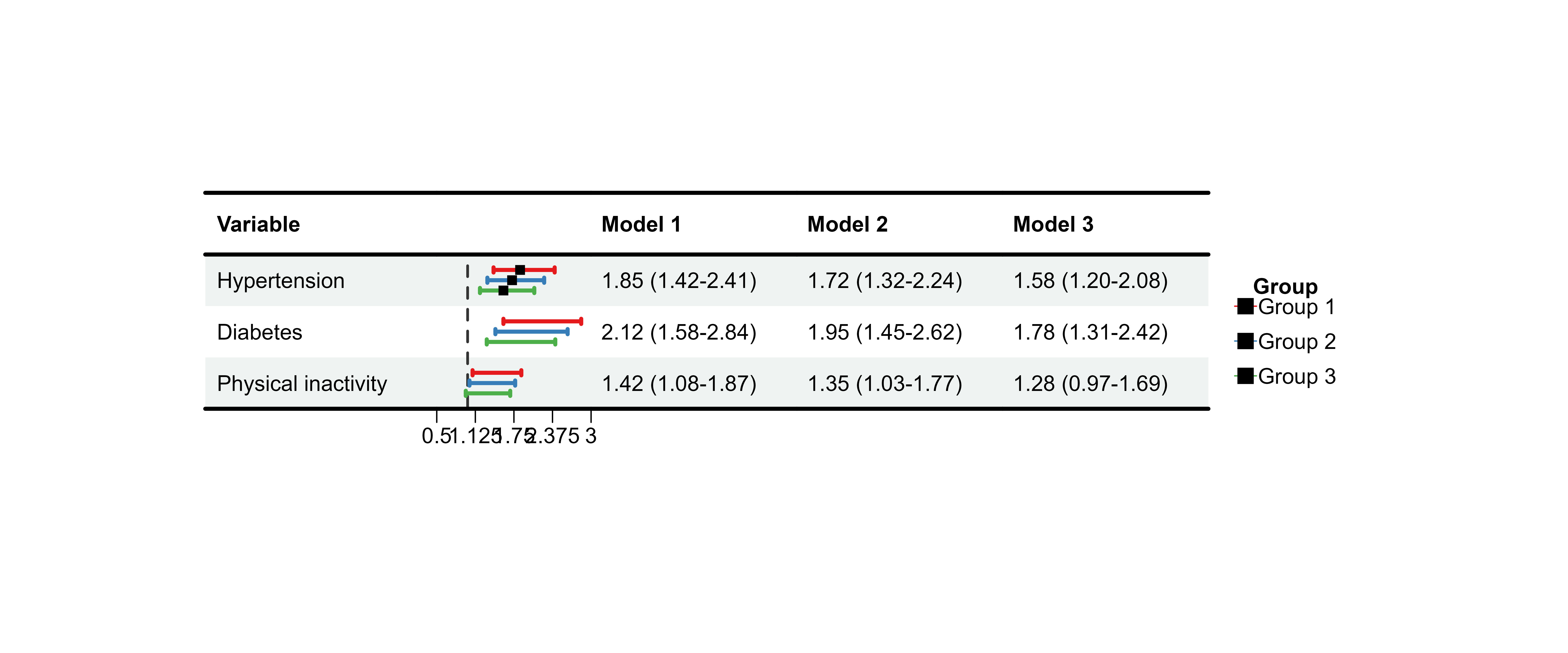
⚠️ Critical: The sizes parameter must
either be: - A single value (applied to all) - A vector matching
nrow(data)
If you provide fewer values, later rows will have no CI displayed!
🎯 Advanced Features
Automatic Tick Generation
When you provide xlim without ticks_at, the
function auto-generates evenly spaced ticks:
p16 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
xlim = c(0.5, 2.5),
ticks_at = NULL # Auto-generate 5 ticks
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 40, 35, 40, 20, 20, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 8
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p16)
Layout Fine-Tuning
Default Layout Parameters
# Default values (can be customized)
# height_top = 8 # Top margin
# height_header = 12 # Header row
# height_main = 10 # Data rows
# height_bottom = 8 # Bottom margin
# width_left = 10 # Left margin
# width_right = 10 # Right marginCustom Layout
p17 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
height_main = 12, # Taller rows
height_bottom = 6, # Smaller bottom margin
width_adjust = 8, # Wider columns
layout_verbose = TRUE # Print layout info
)
#>
#> ── Layout Adjustment Summary
#> ℹ Column Widths (mm):
#> Default: 5, 32.9, 27.5, 34.1, 14.6, 11.1, 5
#> Adjusted: 10, 48, 40, 48, 24, 24, 10
#> width_custom = list({paste(width_code, collapse = ', ')})
#> ℹ Row Heights (mm):
#> Default: 5, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 9.7, 5
#> Adjusted: 8, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 6
#> height_custom = list({paste(height_code, collapse = ', ')})
#> ✔ Tip: Copy and modify the code above for custom dimensions
print(p17)
Manual Override
For pixel-perfect control:
p18 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
height_custom = list('3' = 15, '4' = 15), # Specific rows
width_custom = list('2' = 80, '3' = 100), # Specific columns
layout_verbose = FALSE
)
print(p18)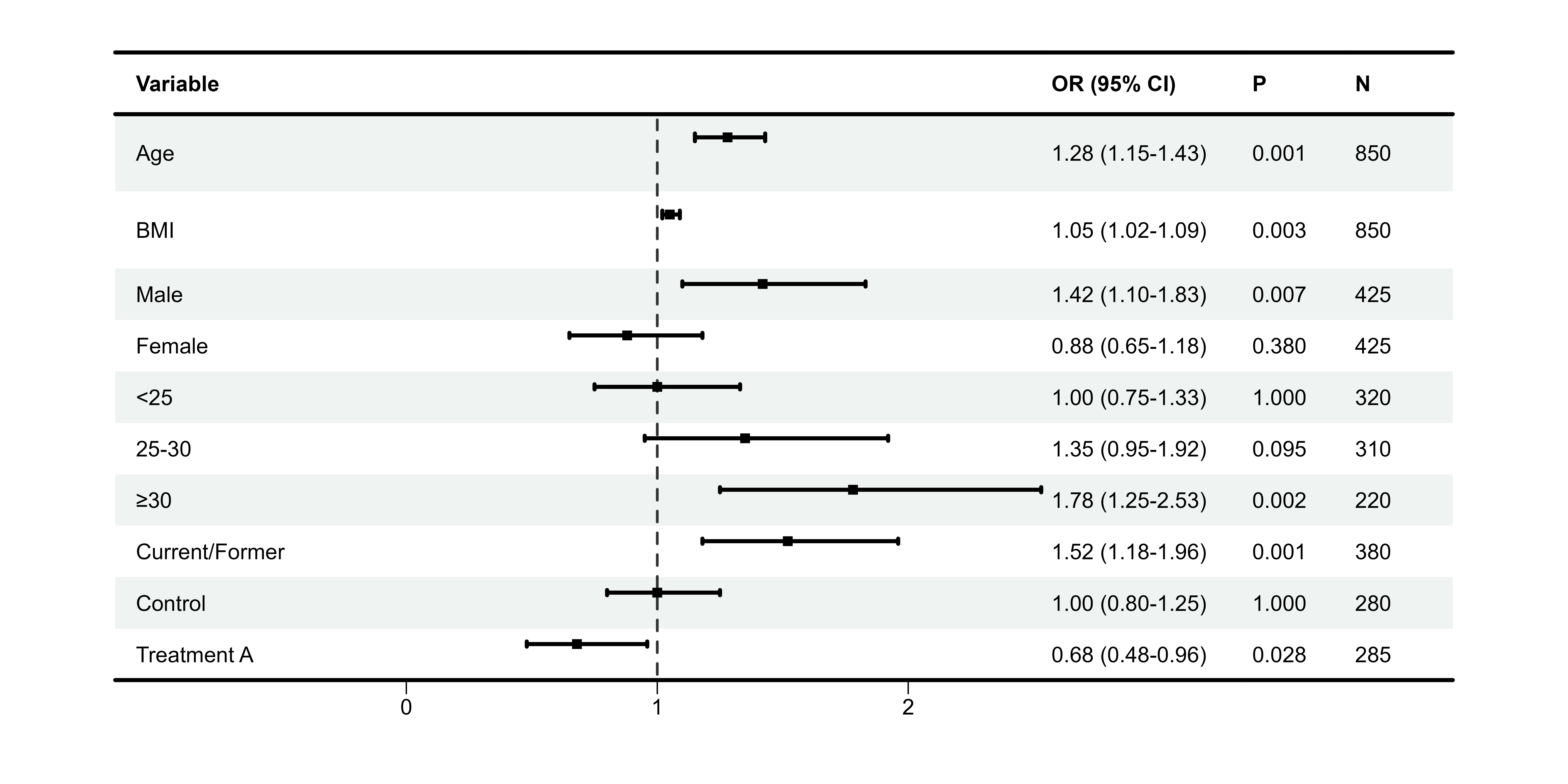
Saving Plots
# Save to multiple formats
p19 <- plot_forest(
data = plot_data,
est = list(df_single$est),
lower = list(df_single$lower),
upper = list(df_single$upper),
ci_column = 2,
ref_line = 1,
save_plot = TRUE,
filename = "my_forest_plot",
save_path = "output",
save_formats = c("png", "pdf", "tiff"),
save_width = 30,
save_height = 25,
save_dpi = 300
)📚 Real-World Examples
Example 1: Logistic Regression Results
# Simulate logistic regression results
set.seed(123)
logistic_results <- data.frame(
Variable = c(
"Demographics", " Age (per 10 years)", " Male sex",
"Clinical", " BMI ≥30", " Hypertension", " Diabetes",
"Laboratory", " CRP >3 mg/L", " LDL-C >130 mg/dL"
),
OR = c(NA, 1.35, 0.82, NA, 1.58, 1.42, 1.67, NA, 1.44, 1.28),
Lower = c(NA, 1.15, 0.65, NA, 1.22, 1.18, 1.32, NA, 1.15, 1.02),
Upper = c(NA, 1.58, 1.03, NA, 2.05, 1.71, 2.11, NA, 1.81, 1.61),
P = c(NA, 0.001, 0.085, NA, 0.001, 0.001, 0.001, NA, 0.002, 0.035)
)
# Prepare display
logistic_display <- logistic_results %>%
mutate(
` ` = strrep(" ", 20),
`OR (95% CI)` = ifelse(is.na(OR), "",
sprintf("%.2f (%.2f-%.2f)", OR, Lower, Upper)),
`P-value` = ifelse(is.na(P), "",
ifelse(P < 0.001, "<0.001", sprintf("%.3f", P)))
) %>%
select(Variable, ` `, `OR (95% CI)`, `P-value`)
# Identify group headers
group_rows <- c(1, 4, 7)
# Create plot
p_logistic <- plot_forest(
data = logistic_display,
est = list(logistic_results$OR),
lower = list(logistic_results$Lower),
upper = list(logistic_results$Upper),
ci_column = 2,
ref_line = 1,
xlim = c(0.5, 2.5),
arrow_lab = c("Protective", "Risk Factor"),
align_left = 1,
align_center = 2,
align_right = c(3, 4),
bold_group = logistic_display$Variable[group_rows],
bold_pvalue_cols = 4,
p_threshold = 0.05,
background_style = "group",
background_group_rows = group_rows,
ci_colors = ifelse(is.na(logistic_results$P) | logistic_results$P >= 0.05,
"#CCCCCC", "#E64B35"),
add_borders = TRUE,
layout_verbose = FALSE
)
print(p_logistic)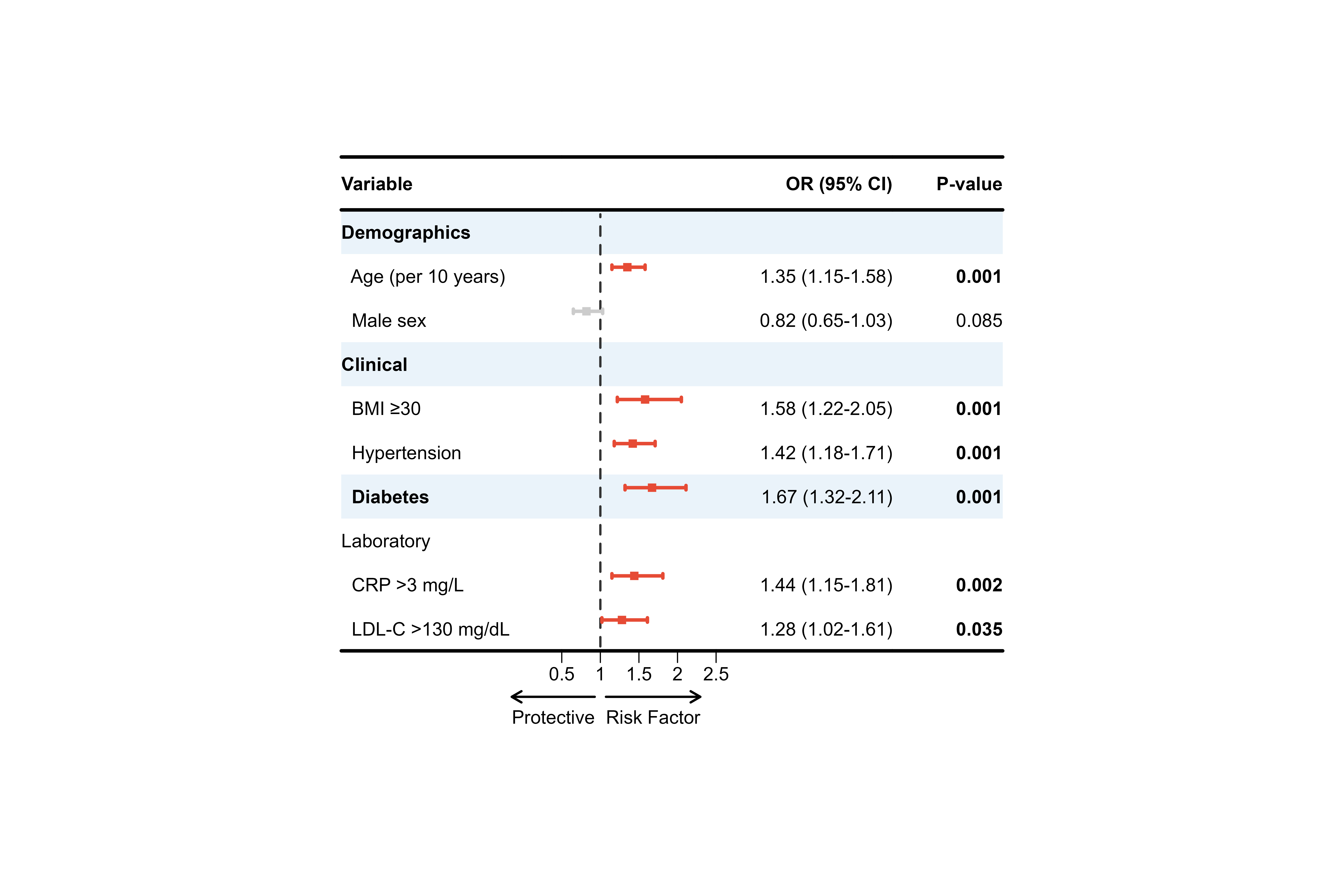
Example 2: Cox Regression (Survival Analysis)
# Survival analysis hazard ratios
cox_results <- data.frame(
Gene = c("BRCA1", "BRCA2", "TP53", "EGFR", "MYC",
"KRAS", "PIK3CA", "AKT1", "PTEN"),
HR = c(1.45, 0.78, 2.12, 1.23, 0.91, 1.87, 1.56, 0.85, 1.34),
Lower = c(1.18, 0.61, 1.58, 0.95, 0.72, 1.42, 1.20, 0.66, 1.05),
Upper = c(1.78, 0.99, 2.84, 1.59, 1.15, 2.46, 2.03, 1.09, 1.71),
P = c(0.001, 0.041, 0.001, 0.124, 0.412, 0.001, 0.001, 0.235, 0.018)
)
cox_display <- cox_results %>%
mutate(
` ` = strrep(" ", 20),
`HR (95% CI)` = sprintf("%.2f (%.2f-%.2f)", HR, Lower, Upper),
`P-value` = ifelse(P < 0.001, "<0.001", sprintf("%.3f", P))
) %>%
select(Gene, ` `, `HR (95% CI)`, `P-value`)
p_cox <- plot_forest(
data = cox_display,
est = list(cox_results$HR),
lower = list(cox_results$Lower),
upper = list(cox_results$Upper),
ci_column = 2,
ref_line = 1,
xlim = c(0.5, 3),
arrow_lab = c("Better Survival", "Worse Survival"),
align_left = 1,
align_right = c(3, 4),
bold_pvalue_cols = 4,
p_threshold = 0.05,
background_style = "zebra",
ci_colors = ifelse(cox_results$P < 0.05, "#E64B35", "#4DBBD5"),
add_borders = TRUE,
height_main = 10,
layout_verbose = FALSE
)
print(p_cox)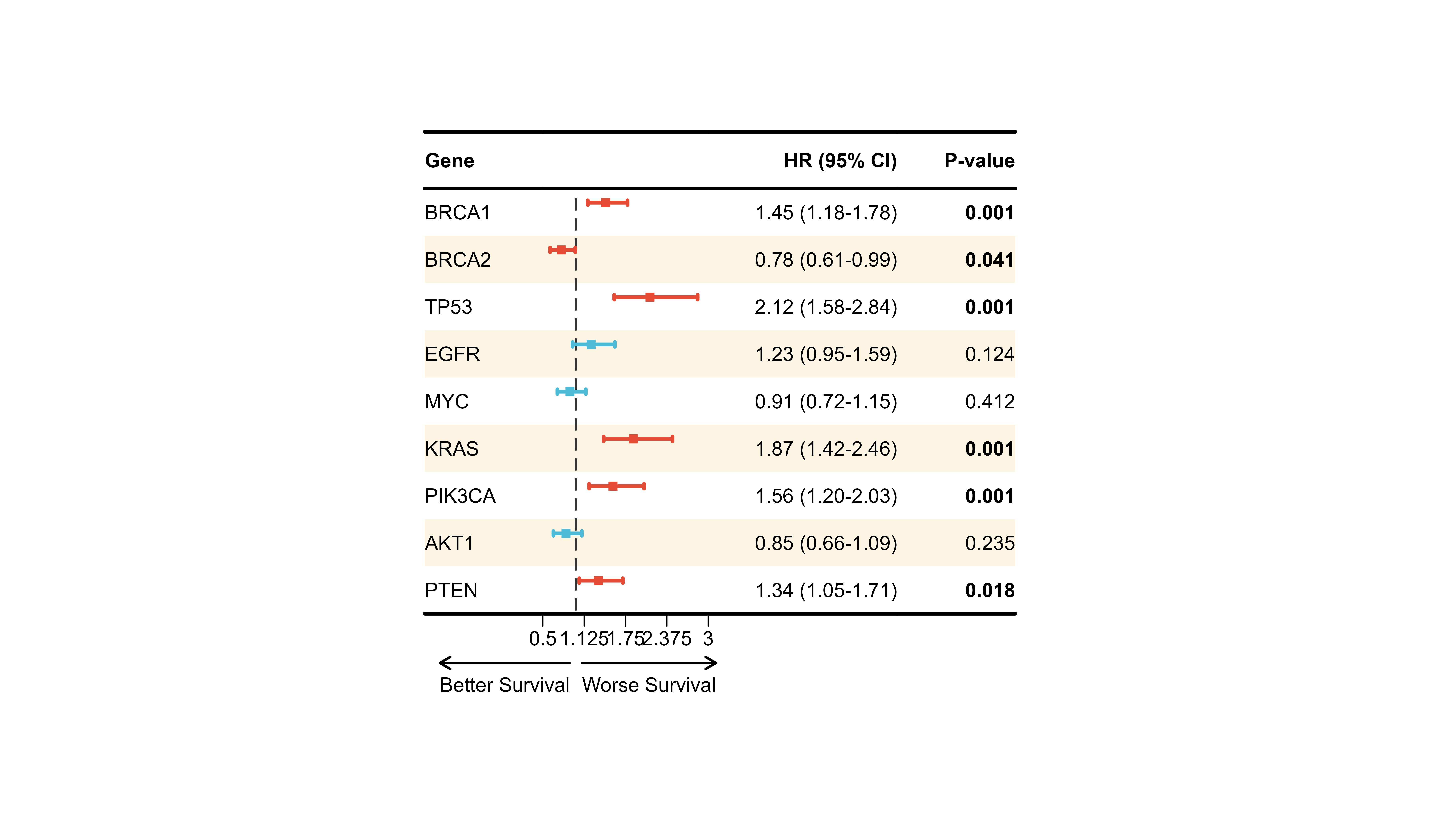
Example 3: Model Comparison (Adjusted vs Unadjusted)
# Use built-in multi-model data
comparison_display <- plot_data_multi %>%
mutate(Note = c(
"Crude model",
"Age + Sex adjusted",
"Fully adjusted"
)) %>%
select(Variable, ` `, `Model 1`, `Model 2`, `Model 3`, Note)
p_comparison <- plot_forest(
data = comparison_display,
est = list(df_multi$est, df_multi$est_2, df_multi$est_3),
lower = list(df_multi$lower, df_multi$lower_2, df_multi$lower_3),
upper = list(df_multi$upper, df_multi$upper_2, df_multi$upper_3),
ci_column = 2,
ref_line = 1,
xlim = c(0.5, 3),
nudge_y = 0.25,
align_left = 1,
align_center = c(3, 4, 5),
align_right = 6,
add_borders = TRUE,
border_width = 4,
layout_verbose = FALSE
)
print(p_comparison)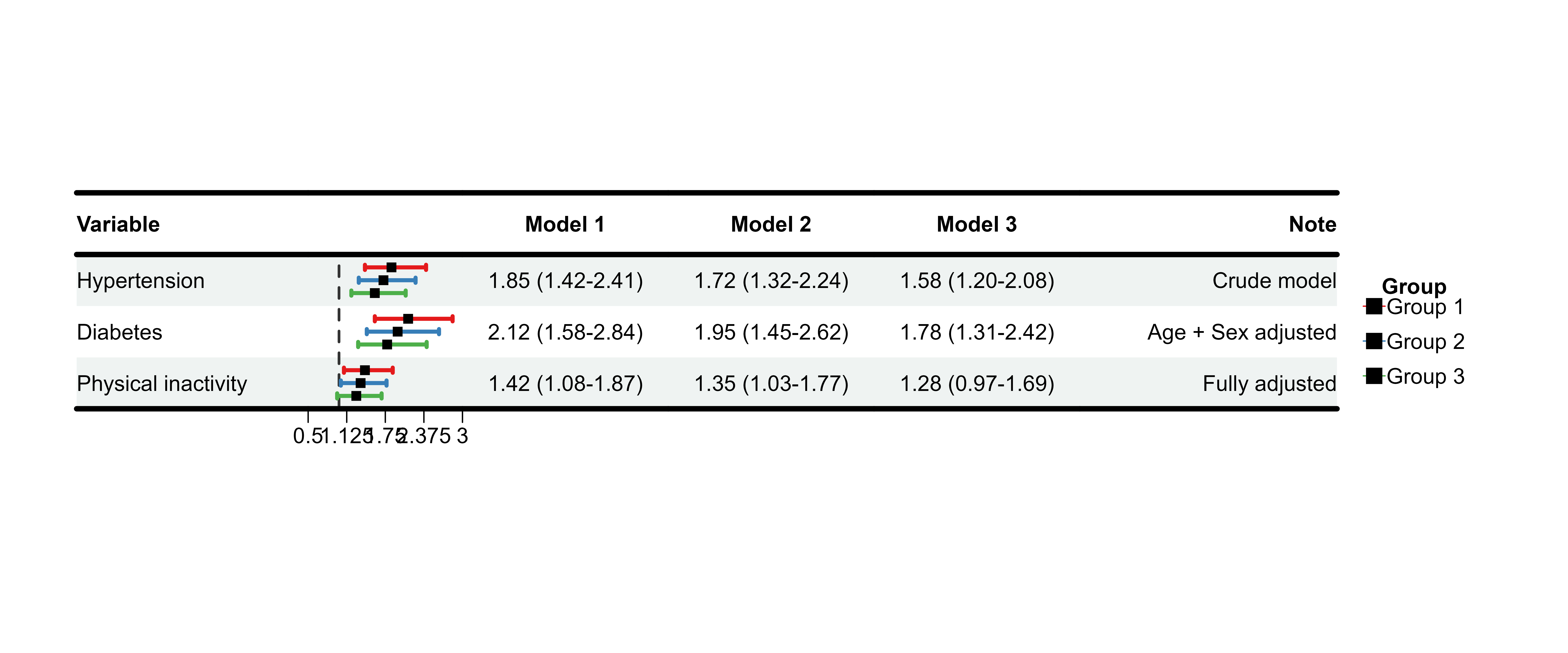
💡 Best Practices
Data Preparation Checklist
✅ Format effect estimates - Use
sprintf() for consistent decimals ✅ Create blank
column - Use strrep(" ", width) for CI graphics ✅
Handle missing values - Use
ifelse(is.na(...), "", ...) for display ✅ Separate
display and numeric - Keep est/lower/upper as separate vectors
✅ Order columns logically - Variable → Blank → Results
→ P-value
Design Guidelines
Color Selection
- Use colorblind-friendly palettes - Test with ColorBrewer or viridis
- Limit colors - 2-3 colors for clarity
- Consistent meaning - Red = risk, Blue = protective, Gray = non-significant
Common Pitfalls
❌ Wrong sizes vector length - Must match
nrow(data), not number of models ❌ Forgetting
blank column - CI graphics need empty space ❌
Inconsistent formatting - Use sprintf()
for uniform decimals ❌ Too many decimals - 2-3 is
usually sufficient ❌ Cluttered axis - Use appropriate
xlim and tick spacing
Workflow Tips
1. Start Simple
# Minimal working example first
p <- plot_forest(data, est, lower, upper, ci_column = 2)2. Add Customizations Incrementally
# Then add features one by one
p <- plot_forest(
...,
align_left = 1, # Step 1
bold_pvalue_cols = 4, # Step 2
background_style = "zebra" # Step 3
)3. Use layout_verbose for Debugging
p <- plot_forest(..., layout_verbose = TRUE)
# Check printed dimensions
# Adjust with height_custom/width_custom if needed4. Save Final Version
p <- plot_forest(...,
save_plot = TRUE,
filename = "final_forest",
save_formats = c("png", "pdf"))📖 Parameter Reference
Essential Parameters
| Parameter | Type | Default | Description |
|---|---|---|---|
data |
data.frame | - | Display data with all text columns |
est |
list | - | Effect estimates (list of vectors) |
lower |
list | - | Lower CI bounds (list of vectors) |
upper |
list | - | Upper CI bounds (list of vectors) |
ci_column |
integer | - | Column index for CI graphics |
ref_line |
numeric | 1 | Reference line position |
Customization Parameters
| Category | Parameters |
|---|---|
| Theme |
theme_preset, theme_custom
|
| Alignment |
align_left, align_center,
align_right
|
| Bold |
bold_group, bold_group_col,
bold_pvalue_cols, p_threshold
|
| Background |
background_style, background_group_rows,
background_colors
|
| Colors |
ci_colors, ci_group_ids
|
| Borders |
add_borders, border_width,
custom_borders
|
| Layout |
height_*, width_*, nudge_y,
sizes
|
| Save |
save_plot, filename,
save_path, save_formats
|
For complete parameter documentation, see
?plot_forest.
🎓 Summary
You’ve learned how to:
✅ Create basic forest plots with plot_forest() ✅
Customize themes, colors, and alignment ✅ Compare multiple models
side-by-side ✅ Apply backgrounds, borders, and formatting ✅ Fine-tune
layouts and save publication-ready figures ✅ Follow best practices for
statistical visualization
Next Steps
- Explore other vignettes - Get Started, Color Palettes
-
Check function documentation -
?plot_forest,?forest_data - Share your plots - Tag us on GitHub with your beautiful forest plots!
📦 Package: evanverse 📧 Questions? Open an issue on GitHub 🌟 Like this package? Give us a star!
Happy plotting! 🌲📊
